Synchronized Histological Image Viewing Architecture
The Synchronized Histological Image Viewing Architecture (SHIVA) is a Java-based visualization and analysis application. SHIVA can process 2D and 3D image files and provides convenient methods for users to overlay multiple datasets.
Visit ForumDownload
Features
- Simultaneous visualization of multiple image volumes.
- Tools for labeling and masking of structures.
- Framework for the Mouse Atlas Project.
Description
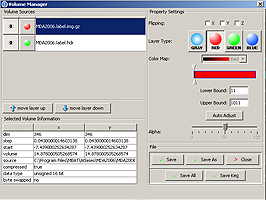
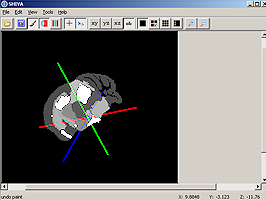
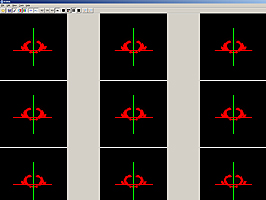
The Synchronized Histological Image Viewing Architecture (SHIVA) is a Java-based visualization and analysis application. SHIVA can load, display, and process 2D and 3D image files. SHIVA provides convenient methods for users to overlay multiple datasets. For example, one can simultaneously display co-registered MRI and histology data with a set of anatomical labels that delineate structures or tissue types. SHIVA currently supports files that are stored in Analyze image, MINC, TIFF, and some vendor specific formats. The distribution of SHIVA includes a Java binary and a collection of data files from the LONI Mouse Atlas Project.System Requirements
Java
- Size: 2.42 Mb
- Memory: 256 Mb
- Software: Java 1.4
Purpose
SHIVA provides a visualization framework that can display simultaneously display multiple image files. SHIVA can display data in single planes, simultaneous orthogonal planes has several isplay modes, including lightbox views, single plane views, or simultaneous orthogonal (axial, sagittal, and coronal) views of 3D image volume datasets. SHIVA also provides tools for delineation of anatomical structures and generation of mask volumes.